Single cell microbial physiology to monitor the water quality in treatment processes and water distribution systems
Drinking water companies are constantly monitoring the water quality in their distribution networks. The companies are mainly interested in quickly detecting deviations in water quality. Whatever the origin of the contamination, rapid identification is needed to ensure water quality and subsequent consumer safety and satisfaction. Despite a wide and ever-increasing variety of molecular techniques available in the microbiologist’s toolbox of analytical methods, their use for the routine microbiological examination of drinking water is still limited and restricted to the presence or absence of (specific) microorganisms. The methods (Q-PCR, PCR, FISH) are mostly used for the detection of specific bacteria (Legionella species) or confirmation of the microbial identity of colonies grown on specific agar plates (coliforms, Clostridia, Aeromonads etc.). Lack of sensitivity, complexity, and the relative costs of molecular, compared to traditional methods and regulatory restrictions, are factors that have limited the acceptance of molecular techniques by water utilities and drinking water testing laboratories. The decrease in costs for equipment, DNA/RNA sequencing, synthesis of DNA probes, and user-friendly bioinformatics software might change this situation rapidly in the future.
Technology
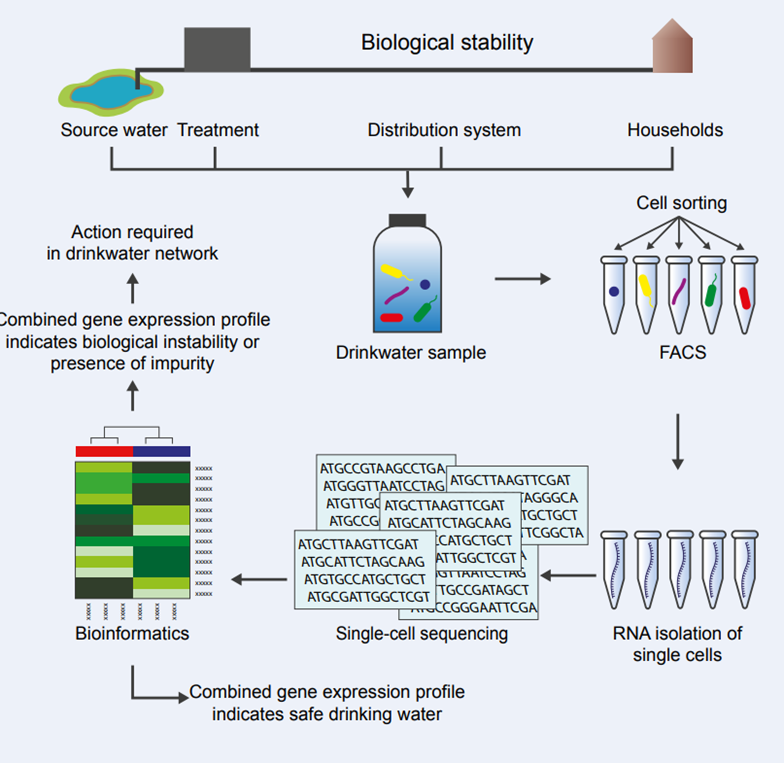
Microbial cells use their environment to translate a part of their DNA into proteins, leading to metabolic activity. RNA is the intermediate between the DNA and the proteins. Sensitive RNA analysis is possible because known technologies are available to amplify the number of RNA molecules to such an extent that allows the detection and identification of the RNA molecule by sequencing. This allows, in principle, the detection of a single RNA molecule in a single microbial cell.
Challenge
The aim of this research is to add an additional advanced molecular biological method to the existing set of analytical methods assessing the chemical and microbial water quality. Potable water quality is determined by standard chemical and microbial analyses using various analytical and growth-based tools. Both methods are based on predetermined assumptions; therefore, current analytical methods might not detect unknown compounds and microorganisms.
Solution
This project will focus on the in-situ quantification of RNA expression patterns at the single-cell level. RNA patterns give information on the microbial community’s metabolic state and indicate the identity and concentration of bioactive substances in the water and which reactions/changes will take place in the water column and associated biofilm.
- Rijksuniversiteit Groningen Vitens Biotrack Evides Vereniging Participanten Waterketen Noord Nederland (WON) Wetsus
Inez Dinkla
Wetsus